 |
|||||||||||
|
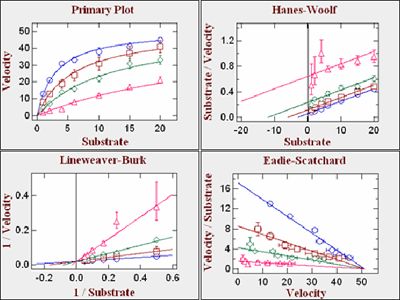 
New in 2010Two Substrate Enzyme Kinetics
|
||||||||||||||||||
| Two Substrate l One Substrate One Activator l Tight Binding Kinetics l Equilibrium Binding l Binding Calculator | |
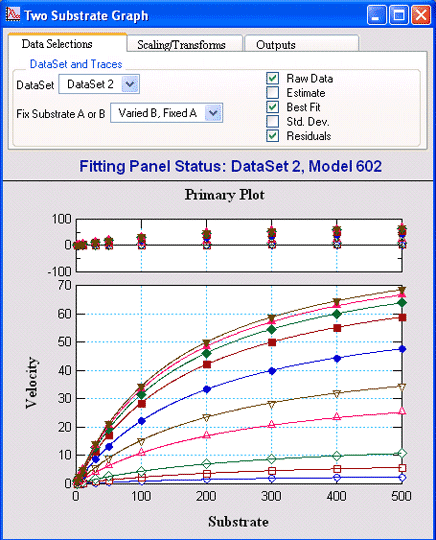 Two Substrate Kinetic DataVisualEnzymics provides seven equations for
fitting two substrate steady state rate saturation
profiles. These include Ordered Bi Uni, Random Bi
Bi, Random Bi Bi with Product Inhibition, Random Bi
Bi with Substrate Inhibition, Ping Pong, Ping Pong
with Competitive Substrate Inhibition, and Ping Pong
with Double Competitive Substrate Inhibition. |
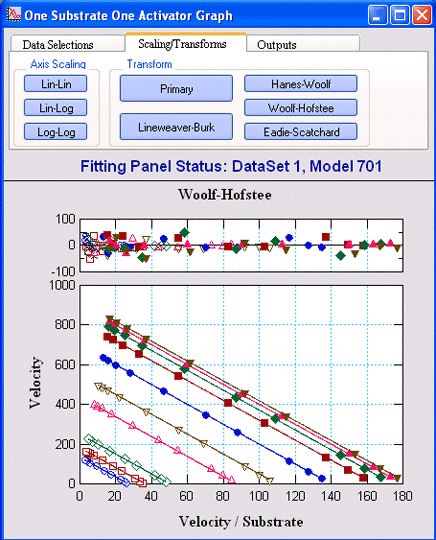
One Substrate One ActivatorVisualEnzymics provides eight
equations for fitting enzyme reactions that display
essential and non-essential activation by metal ions
or ligands. These include reactions where the
activator binds to the substrate and the true
substrate is the substrate-activator complex, and
reactions where the activator binds to the enzyme in
an activator site. |
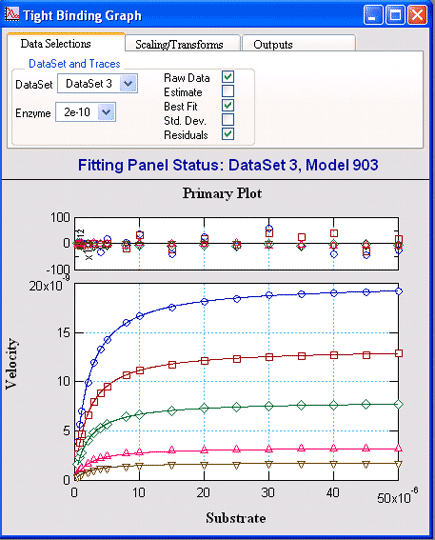 Tight Binding KineticsVisualEnzymics provides four equations for tight
binding competitive, uncompetitive, noncompetitive,
and mixed inhibition. These equations apply to
reactions where the inhibitor Kd approximates the
enzyme concentration, and where the true kinetic
equation includes a quadratic term to correct for
inhibitor depletion due to binding. |
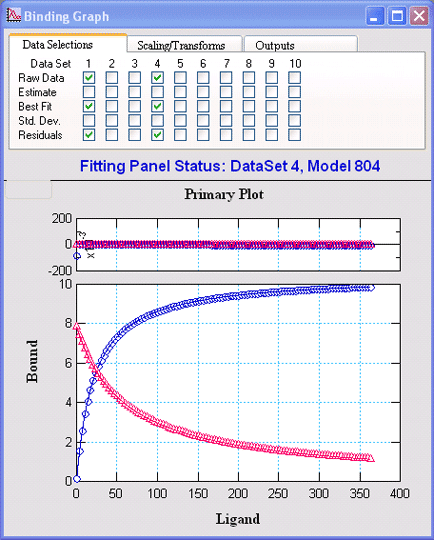
Equilibrium BindingVisualEnzymics provides eight
equations for analysis of ligand binding to one or
two sites, as well as competitive binding reactions
to one site. The equations provide quadratic and
cubic solutions to binding equilibria under
conditions where all the ligands are present at
similar concentrations, and the equations fit for
the true binding constants. |
|
|
|
|
Visual Enzymics Improvements in 2008 Data Masking l Parallel Processing l Igor Pro 6 |
|
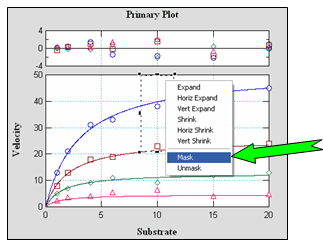 |
Data MaskingVisualEnzymics gives the user the option to include or exclude data points in curve fits by selecting data in the graph window. Points can be masked or unmasked with the marquee and popup menu. More than one point can be masked from different parts of the same curve, or multiple points on separate data curves. Visually, masked points are dimmed in the graph window to indicate that they are not included in the current fit. Statistical reports indicate which points have been excluded in the reported results. Data masking can be used to identify regions of the curve
fit that are most sensitive to data variation. By
comparing curve fits that include or exclude
specific data, new experiments can be designed that
incorporate more data in the sensitive regions. More
data and better experimental design provide higher
confidence in discriminating between competing
kinetic models. |
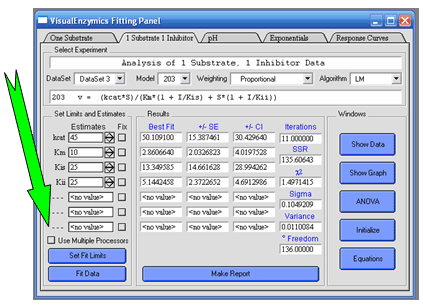
Parallel ProcessingVisualEnzymics now uses Igor Pro's built in support for multi-processor curve fitting. Select the multi-processor option to generate independent computational threads on each core. The software will automatically detect the number of processors in the computer and then generate as many computational threads as there are processors. Parallel processing can speed up computation when fitting large data sets to complex kinetic models. |
 |
 |
Igor Pro 6VisualEnzymics supports all the functionality of the latest Igor Pro release. Igor Pro 6 on Macintosh now runs natively on either Intel or Power PC processors. New features in Igor Pro 6 include a statistics package with >150 new capabilities for analyzing data, 33 example experiments in the statistics folder, multi-processor capabilities for curve fitting, quick fit menu to skip the standard curve fitting dialog, curve fitting with orthogonal distance regression algorithm, automatic annotation of fit results added to graphs, improved drawing capabilities in layouts, panels, and graphs, external tool palettes for graphics windows, new graph Fling feature to slide graph contents in all directions, improved table formatting and functionality, magnifiers for all text windows, new action commands from notebook text links, improved listbox controls for creating complex listboxes that display graphs and images, floating panel capabilities, exterior subwindow panels, new matrix math operations………. and many more. If you analyze scientific data, Igor Pro is the top choice for Macs and PCs.
|
| Softzymics, Inc. 113 Scarborough Place, Charlottesville, VA 22903, Phone 434-422-0446. Copyright 2002-2016, Softzymics, Inc. All Rights Reserved
|